Software
- To achieve our lab’s central goal and identify mechanisms of aberrant gene expression in cancer, we also innovated software and algorithms to dissect cellular and molecular heterogeneity. These tools have enabled several basic and translational studies in cancer, as well as diabetes and Alzheimer.
AnnoSpat
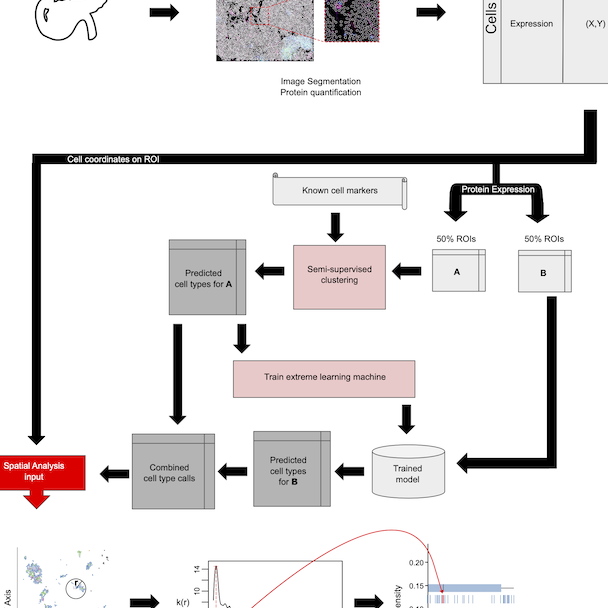
A computational tool for automated cell type prediction and cell neighborhood quantification for Image Mass Cytometry (IMC) and Co-Detection by Indexing (CODEX).
AnnoSpat annotates cell types and quantifies cellular arrangements from spatial proteomics
Mongia A, Zohora FT, Burget NG, Zhou Y, Saunders DC, Wang YJ, Brissova M, Powers AC, Kaestner KH, Vahedi G, Naji A, Schwartz GW, Faryabi RB Nature Communications, 2024
PubMed PMID: 38702321;
PMCID: PMC11068798

PANC-DB
PANC-DB is a database and web portal for dissemination and visualization of Human Pancreas Analysis Program’s cellular and molecular datasets.
Multiomics Single-cell Analysis Of Human Pancreatic Islets Reveals Novel Cellular States In Health and Type 1 Diabetes.
Nature Metabolism
PubMed PMID: 35228745;
PMCID: PMC8938904
TooManyPeaks
New addition to our graph-based single-cell analysis environment that enables cell type prediction and visualization from single-cell ATAC-seq data.
TooManyPeaks Identifies Drug-resistant-specific Regulatory Elements From Single-cell Leukemic Epigenomes.
Schwartz GW, Zhou Y, Petrovic J, Fasolino M, Xu L, Pear WS, Faryabi RB Cell Reports, 2021
PubMed PMID: 34433064;
PMCID: PMC7439807

TooManyCells
A suite of graph-based tools for efficient, global, and unbiased identification and visualization of single-cell RNA-seq clustering.
TooManyCells Identifies And Visualizes Relationships Of Single-cell Clades.
Schwartz GW, Zhou Y, Petrovic J, Fasolino M, Xu L, Shaffer SM, Pear WS, Vahedi G, Faryabi RB Nature Methods, 2020; 17: 405-413
PubMed PMID: 32123397;
PMCID: PMC7439807
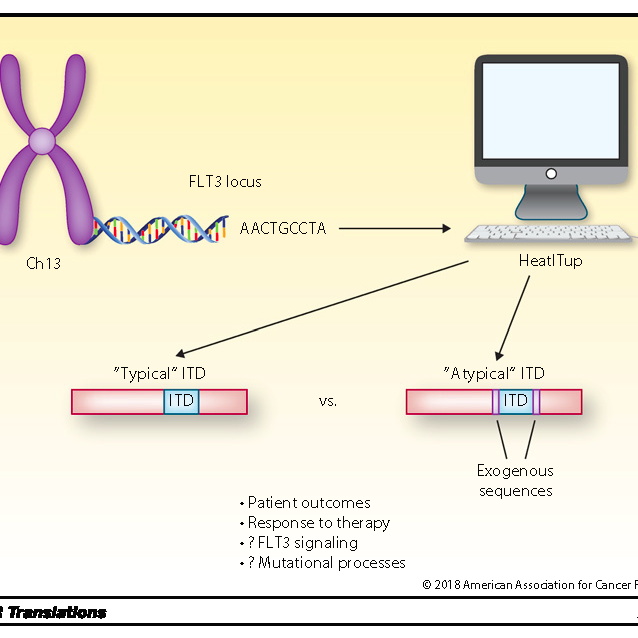
HeatITup
Internal tandem duplication (ITD) mutations are diverse and complex genetic alterations, commonly observed in many cancers including 60% of acute myeloid leukemia. HEAT diffusion for Internal Tandem dUPlication (HeatITup) provides an efficient and robust algorithm for identification, classification, and visualization of ITD mutations.
Classes of ITD Predict Outcomes in AML Patients Treated With FLT3 Inhibitors.
Schwartz GW, Manning BS, Zhou Y, Velu PD, Bigdeli A, Astles R, Lehman AW, Morrissette JJ, Perl AE, Li M, Carroll M, Faryabi RB Clinical Cancer Research 25(2): 573-583, Jan 2019
PubMed PMID: 30181385;
PMCID: PMC7439807
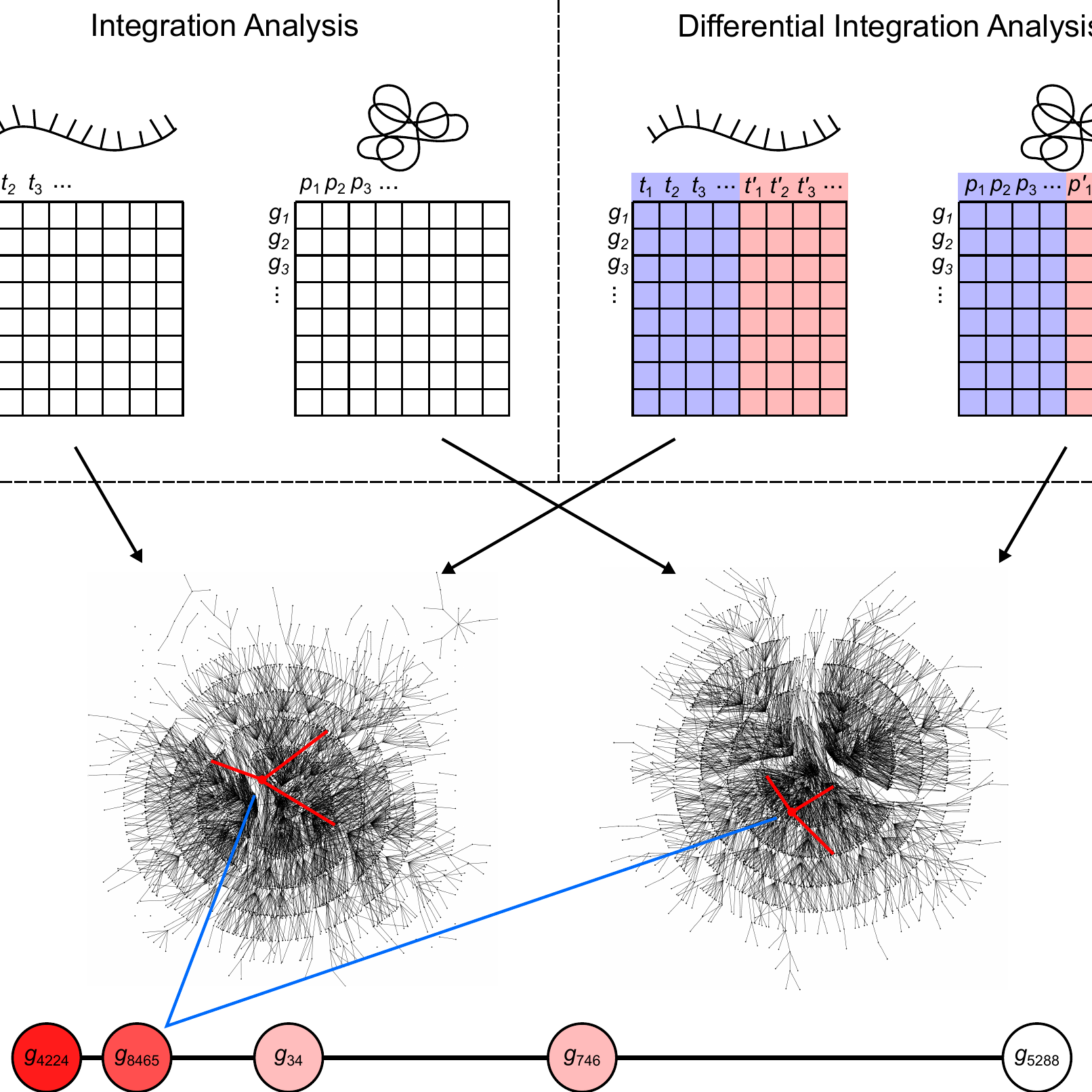
InteGREAT
Transcriptomic and proteomic measurements provide complementary but sometimes irreconcilable information about tumors. To accurately relate heterogenous high-throughput data sources, we developed inteGREAT, a graph-based algorithm. inteGREAT provides a robust and scalable solution for differential integration of transcriptomic and proteomic measurements. Application of inteGREAT to breast cancer data identified novel prognostic biomarkers associated with different disease subtypes.
Differential Integration of Transcriptome and Proteome Identifies Pan-cancer Prognostic Biomarkers.
Schwartz G, Petrovic J, Zhou Y, Faryabi RB Frontiers in Genetics 9: 205, June 2018
PubMed PMID: 29971090;
PMCID: PMC6018483